Nucleic acid detection/staining
CHOOSE THE RIGHT STAIN FOR YOUR APPLICATION
| Product | Method | Procedure | Advantages | Disadvantages | Recommended for |
|---|---|---|---|---|---|
| DNA prestaining | GelRed® Prestain Plus 6X DNA Loading Dye | GelRed® loading buffer is added directly to the DNA sample before loading |
|
|
|
| Precast staining | GelRed® 10,000X in water or GelGreen® 10,000X in water | GelRed® or GelGreen® is mixed with molten agarose before gel casting | Familiar protocol, rapid results | ||
| Precast staining | GelRed® Agarose LE or GelGreen® Agarose LE | Agarose is supplied pre-coated with GelRed® or GelGreen®, just dissolve, heat, and pour | Safer & more convenient, no need to handle concentrated dye | ||
| Post-electrophoresis staining | GelRed® 10,000X in water or GelGreen® 10,000X in water – or – GelRed® 3X in water | No fluorescent dye is added to the gel, it is stained in 3X GelRed® or 3X GelGreen® solution after electrophoresis |
|
Extra staining step (up to 30 minutes) after electrophoresis (some customers report good results after only 5 minutes if dye is not reused) |
|
| Post-electrophoresis staining of PAGE Gels | PAGE GelRed® 10,000X or 1X in water | No fluorescent dye is added to the gel, it is stained in 1X PAGE GelRed® solution after electrophoresis |
|
Extra staining step of approx. 30 minutes after electrophoresis | Staining of nucleic acids in PAGE gels |
GelRed™ and GelGreen™ - DNA Stains Biotium
Convenient for the Lab – environmentally friendly
To visualize nucleic acids the experimenter makes use of color stains that interact with them. One of those stains is ethidium bromide (EtBr) which can be illustrated in an agarose or polyacrylamide gel when exposed to UV rays. EtBr is proven to be a highly toxic, cancerous and mutagenic substance which calls for cautious handling. It is linked to frameshifting in the DNA which leads to mutation and therefore a loss of function in proteins. To minimize factors of possible damages that are accompanied with the use of EtBr other substances like SYBR Green I + II, OliGreen or RedSafe which are said to be less harmful can be used.
Biotium offers two alternatives to EtBr which do not penetrate neither latex gloves nor the cell membrane and therefore cannot interact with the DNA - GelRed™ and GelGreen™. Additionally a higher sensitivity for better quantification and less sample use can be achieved.
Mechanism of DNA Intercalation of ethidium bromide
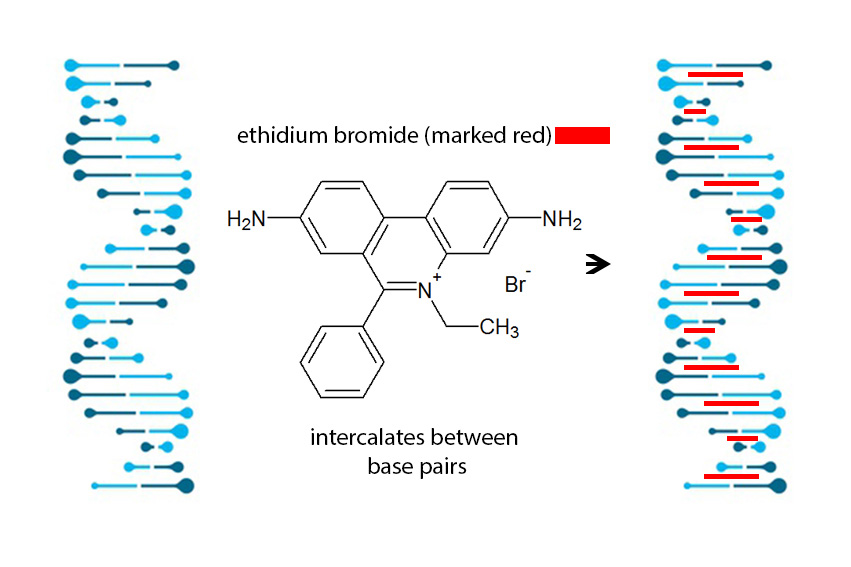
Figure 1 ethidium bromide (red bars) settles between base pairs and alternates the structure. This leads to a shift in reading frame of the DNA. For visualization of DNA essential. Instead of EtBr you can use GelRed™ and GelGreen™.
Differences of GelRed™ and GelGreen™ to common stains (ethidium bromide, SYBR)
1. Do not penetrate Latex gloves.
a. Glove Penetration Test
2. Do not penetrate cell membrane.
a. Cell staining test
3.Biodegradable.
a. Can be disposed down the drain or in regular trash.
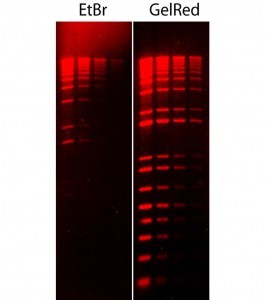
Figure 2 Comparison of ethidium bromide (EtBr) and GelRed in precast gel staining in 1% agarose/TBE gel. Two-fold serial dilutions of 1 kb Plus DNA Ladder (Invitrogen) were loaded in the amounts of 200 ng, 100 ng, 50 ng and 25 ng from left to right. Gels were imaged using 300 nm transilluminator and photographed with an EtBr filter.
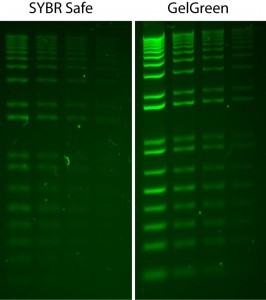
Figure 3 Comparison of GelGreen and SYBR Safe in post-electrophoresis staining of 1% agarose/TBE gels. Two-fold serial dilutions of 1 kb Plus DNA Ladder (Invitrogen) were loaded in the amounts of 200 ng, 100 ng, 50 ng and 25 ng from left to right. Gels were imaged using 254-nm transilluminator and photographed with a SYBR filter.
For more information read the Safety Report of Biotium.
Literature:
(1) Mülhardt, Der Experimentator – Molekularbiologie Genomics, K.3 S. 62
Image register
(1) Mechanism of ethidium bromide intercalation into DNA. Created with ChemSketch and Photoshop
(2) https://biotium.com/wp-content/uploads/2013/09/41003-EtBr-vs-GelRed-labeled-wider-266x300.jpg Status: 29.03.2016 15:25
(3) https://biotium.com/wp-content/uploads/2013/09/41005-SYBRSafe-vs-GelGreen-labeled-266x300.jpg Status: 29.03.2016 15:25



